
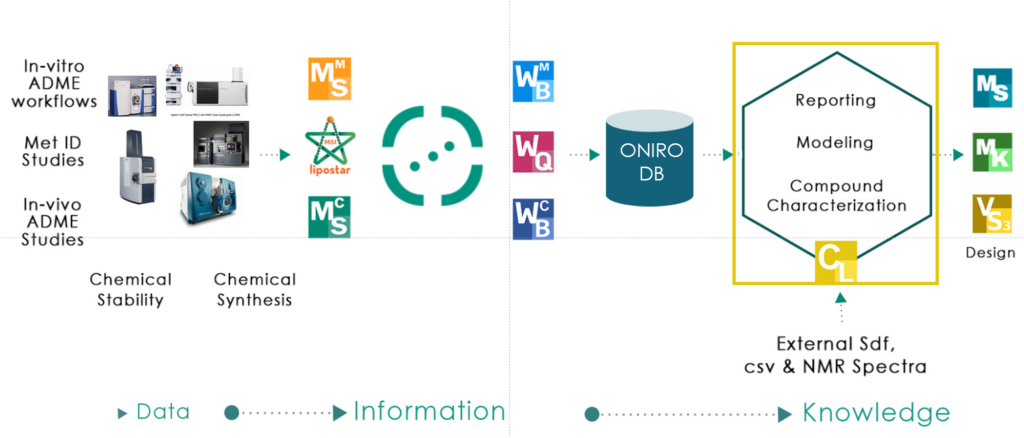
Compound library is a web application that is connected to a database of chemical structures where users can perform multiple computations like NMR prediction but also NMR data spectra interpretation. The information for this database can come from an external sdf file and/or the experiments processed in Web-Quant or WebChembase or WebMetabase within Oniro. Compound Library is distributed as a module of the Oniro system. The system can related the information from drug metabolism studies (met ID or clearance) from multiple experimental protocols. The system has also access to multiple prediction tools like MetaSite, VolSurf, Moka or Charming that covers a wide variety of properties like: Site of Metabolism, molecular descriptors, ADME prediction (permeability, clearance, solubility, protein binding, etc), physico-chemical descriptors (pka, log D, log P) and NMR chemical shift and multiplicity. In addition the system also contain information about the spectrometry of the compounds for both mass spectrometry (MS) and nuclear magnetic resonance (NMR). Finally, the compound library has also access to multiple reporting tools that allows to combine in a single report the results from experiments, in-silico or imported information. The database system is chemical enabled, which mean that the system can perform sub structural search. Also the system can work with small and macromolecules.
If you want to know more about our solutions, do not hesitate to visit our recorded videos of our Practical Applications for Drug Discovery 2020 Webinars.
New features of Compound Library 2.0:
- Data import: The data can be imported from a sd file or comma separated file. The properties of the compounds are mapped to the existing properties in the database giving the option to the end user to modify the mapping done.
- Modeling tool: The user can import/draw any molecule aand perform computations using MetaSite, Moke, MetaDesign or VolSurf on the compound and then decide if it is worthy to keep the compound in the database using a look up system the system also check if the compounds was already existing in the database.
- Compound property: The system it is highly flexible to accommodate computed properties, imported properties or properties generated from WebMetabase and/or WebQuant. Also the system offers the possibility to aggregate the data in a single visualization for the compound.
- Search engine: The system offers the possibility to search the compounds in the database by compound, substructure of any other property with the possibility to mix different search criteria .
- Compound set definition: The suer can after performing a search keep the compounds that were matching the search criteri ina compound set that can then be used to perform computations or aggregation of experiment/computed properties for the entire set, with the possibility to visualize the results in a excel type of sheet that can be also exported.



