The site of metabolism prediction software
MetaSite is a computational procedure that predicts metabolic transformations related to cytochrome and flavin-containing monooxygenase mediated reactions in phase I metabolism. The MetaSite algorithm is unique in being the only software that is not training set dependent, and therefore exhibits improved predictive performance for novel pharmaceutical compounds.
MetaSite considers both enzyme-substrate recognition, which is a thermodynamic factor, and the chemical transformations induced by the enzyme, which is a kinetic factor.
The MetaSite flowchart was recently improved by experimental information from the Human CYP Consortium Initiative, a joint venture between pharmaceutical companies and Molecular Discovery, working together to solve the most important issues in drug metabolism (for further information on CYP Consortium Initiative, see this page). MetaSite gives unprecedented prediction performance, and has now been updated to include the major FMO isoform (FMO3). Key features include the automatic suggestion of fragment modification to optimize specific metabolic issues (MetaDesign), and an interaction map of the substrate with the enzyme cavity (32D) to aid optimisation in the context of the enzyme.
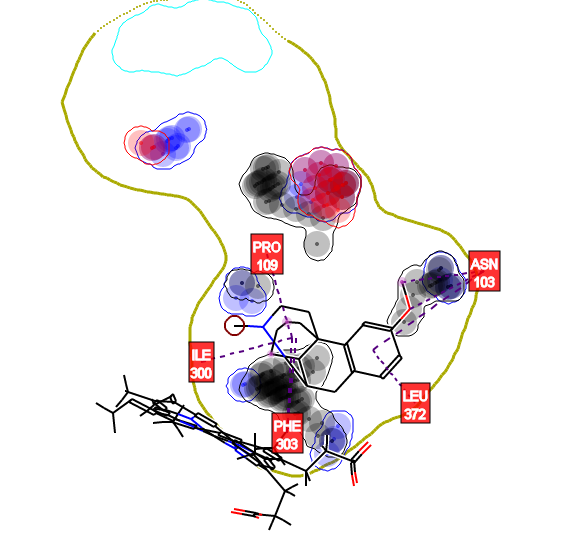
Validation of MetaSite in CYP-Consortium data and in more than forty pharmaceutical companies has shown that the primary site of metabolism was found within the top three MetaSite predictions for more than 90% of the cases.
In addition, MetaSite provides the structure of the metabolites formed with a ranking derived from the site of metabolism predictions. The method also highlights the molecular moieties that help to direct the molecule in the cytochrome cavity such that the site of metabolism is in proximity to the catalytic centre.
Directly blocking the primary site of metabolism can risk creating an inhibitor of the cytochrome, or may negatively affect the activity or selectivity of the compound towards its therapeutic target. Modifying these contributing moieties that most influence the site of metabolism can bypass both of these potential problems.
To request a copy of Metasite, visit Molecular Discovery site.
Features in Metasite 6.0
- Predict “hot spots” in the molecule to help chemists focus their design of compounds to reduce CYP and FMO3 mediated metabolism.
- Suggest the regions that contribute most towards each “hot spot”, providing additional derivation sites for chemists to design new stable compounds.
- Predict the structures of the most likely metabolites, with exact mass and relative retention time to help and complement the experimental elucidation of metabolite structures.
- Warn about the potential of CYP mechanism-based inhibition, one of the major metabolic hurdles in the development of new safer drugs.
- Produce Phase I and Phase II metabolite structures to be automatically used in Mass-MetaSite software.
- Substrate-specificity and catalytic-activity model for FMO3
- 32D view of FMO3 site and substrate in terms of energetic interactions.
- Human AOX isoform selectivity and SoM models.
- Electrophilicity calculation.
References
- From Experiments to a Fast Easy-to-Use Computational Methodology to Predict Human Aldehyde Oxidase Selectivity and Metabolic Reactions Cruciani G, Milani N, Benedetti P, Lepri S, Cesarini L, Baroni M, Spyakis F, Tortorella S, Mosconi E, Goracci L, J. Med. Chem., 2018, 61 (1), pp 360–371
- Flavin Monooxygenase Metabolism: Why Medicinal Chemists Should Matter. Cruciani G, Valeri A, Goracci, L, Pellegrino RM, Buonerba F, Baroni M. J. Med Chem., 2014.
- Exposition and reactivity optimization to predict sites of metabolism in chemicals. Cruciani G, Baroni M, Benedetti P, Goracci L, Fortuna CG Drug Discovery Today: Technologies 2013, 10(1):e155–e165.
- CYP2C9 Structure−Metabolism Relationships: Optimizing the Metabolic Stability of COX-2 Inhibitors.Ahlström MM, Ridderström M, Zamora I, Luthman K. J Med Chem., 2007 Sep 6;50(18):4444-52.
- Comparison of methods for the predictions of the metabolic sites for CYP3A4-mediated metabolic reactions.Zhou D, Afzelius L, Grimm SW, Andersson TB, Zauhar RJ, Zamora I. Drug Metab Dispos., 2006 Jun; 34(6):976-83.
- MetaSite: Understanding Metabolism in Human Cytochromes from the Perspective of the Chemist. Cruciani G, Carosati E, De Boeck B, Ethirajulu K, Mackie C, Howe T, Vianello R. J Med Chem., 2005 Nov 3;48(22):6970-9.



