
More than just a metabolite database
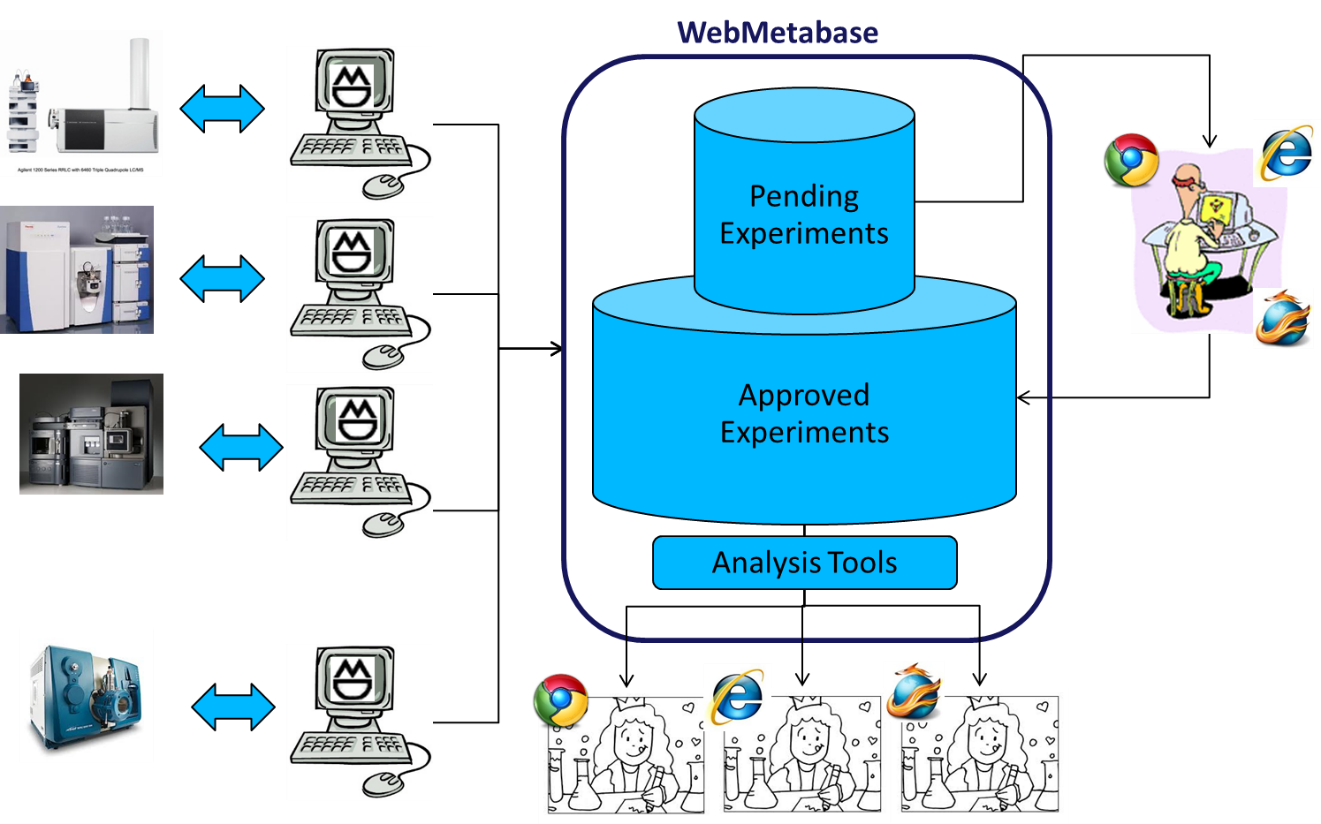
WebMetabase is a server-based application that is used for metabolite identification data storage, reviewing metabolite identification experiments, and extracting the maximum knowledge from the information loaded into the system. In combination with MassMetaSite and after the approval of a Met ID expert, WebMetabase speeds up the process of translating the spectral information into chemical structures, transforming the instrument data into information.
The information can then be used with quantitative and qualitative approaches to extract knowledge about clearance or half-life (in the case where incubation kinetics are used) or matrix selectivity when different matrices are analyzed in the experiment. Moreover, MetaDesign has been added as a tool to automatically suggest chemical modifications to improve metabolic properties. In addition to all of these tools to transform information into knowledge based on a single compound analysis, WebMetabase offers innovative approaches to consider in a single view the metabolic pathways for a number of closely related series of compounds, the Structure Metabolism Relationship Table (SMrT).
Due to the high flexibility of the system, it can accommodate a number of ADME dedicated workflows, among them: Soft Spot Analysis, Glutathione reactive metabolite trapping, cytochrome reaction phenotyping, species comparison, etc. All the workflows could be computed for both small and peptide molecules.
New features in WebMetabase 4.1
- Workflow automation: The system is enabled to start the workflow from a sample list and it will automatically generate the protocol (if needed) and the experiments, and also if set up it will run MassMetaSite within WebMetabase. Therefore, the user will only need to import the sample list, have/copy the samples in the right folder or UNIFI analysis and wait for the results to show up in the WebMetabase interface.
- Excel report: The user can export the information in the chromatogram tab in a excel table with a right click on the table.
- Metabolite notes: The user can add notes to each metabolites in addition an over all note to the experiments
- Fragments reported: The user can have the visualization of all the fragments generated for a metabolite in a single screen and select the ones to output in a report.
- Gal filler: The user can use this functionality in order to automatically search using a extracted ion chromatogram for every peak that is found in a sample into the other samples of the same experiment, ensuring in this way that now peak is missed in the interpretation of the data. All of this integrated in a automation protocol that can remove/hide peaks before this operation depending on multiple customized filter parameters.
- MassMetaSite 4.1 new columns integration: The new columns generated in MassMetaSite 4.1 with respect to the Area ratio between the sample under consideration and the blank sample is introduced, also the column that control the quality of isotope pattern is also imported.
Other features:
- Flexible experimental protocol definition (properties, macros, filters…).
- Automatic uploading of Mass-MetaSite or Metabolite Pilot experiments.
- Treatment of experiments with several samples, i.e. multiple incubation times, matrices, species.
- Expert user for experiment approval and control of the information shared with the rest of the organization.
- Chromatogram browser:
- Support for UV and radio label chromatograms.
- Filter of the solutions Extracted Ion Chromatogram by the existence of a peak above a user specified area.
- Easy data sharing system.
- Flexible reporting of data with customizable reports.
- Analysis tools. Simple compound toolbox:
-
-
- MetaDesign
- Kinetic analysis for parent and metabolites
- Quan/Qual approach
- Three2D: Visualization of the interaction of the parent compounds with different cytochrome enzymes considering the structure of the metabolite as starting point
- Metabolic pathway analysis: The user can easily draw a metabolic pathway keeping the relationship between the different metabolites
- Reporting system: A totally flexible report system has been implemented
- Analysis tools. Multiple compound toolbox:
- Structure Metabolism relationship Table
- Clustering View: This analysis tools enables the comparison of the experimental results for the same compound in different experimental setups
- The comparator, a tool that compares the metabolite structures from different experiments even if the Markush structures are not matching 100%).
- The fragment analysis that compute the frequency of occurrence for a particular reaction in a specific chemical moiety.
- Sdf export: All the metabolites can be exported in sdf format.
-
To request a copy of MassMetasite, visit Molecular Discovery site.



